The life sciences encompass a vast and dynamic field that seeks to unravel the mysteries of living organisms. To address the complexities of biological systems, researchers in the life sciences employ a diverse range of research strategies. These strategies are designed to investigate fundamental questions, elucidate molecular mechanisms, and ultimately contribute to advancements in medicine, agriculture, and environmental science. This essay explores key research strategies in the life sciences, highlighting their significance and providing references to support their application.
- Experimental Design and Hypothesis Testing:A cornerstone of life sciences research is the development of well-designed experiments and the formulation of testable hypotheses. Researchers systematically manipulate variables to observe their effects, providing a foundation for evidence-based conclusions. This approach is exemplified in studies such as those by Silverman and Deitcher (2013) on synaptic transmission in Drosophila, where meticulous experimental design contributed to the understanding of neurotransmitter release.
Experimental design and hypothesis testing are fundamental components of the scientific method, providing a structured approach to investigating and understanding natural phenomena. In the life sciences, these methodologies are crucial for formulating and validating theories, elucidating biological mechanisms, and advancing scientific knowledge. This comprehensive overview explores the key aspects of experimental design and hypothesis testing, delving into their significance and practical application.

1. Formulating a Hypothesis:
At the core of the scientific process lies the formulation of a hypothesis—a clear and testable statement predicting the outcome of an experiment. The hypothesis is often derived from prior observations, existing theories, or gaps in current understanding. For example, a researcher studying the effects of a new drug on a specific cellular process may hypothesize that the drug will enhance or inhibit the observed activity based on its known mechanisms of action.
2. Variables and Controls:
Experimental design involves identifying and manipulating variables to assess their impact on the observed phenomenon. Variables can be classified into independent (manipulated), dependent (measured), and controlled variables. Controlling extraneous variables ensures that observed effects can be confidently attributed to the manipulated variable rather than external factors. For instance, in a study on plant growth, variables such as light intensity, soil composition, and temperature would need careful control.
3. Randomization and Replication:
Randomization is a key element in experimental design, helping to minimize bias and ensure that treatment groups are comparable. Random assignment of subjects or samples to experimental conditions reduces the likelihood of confounding variables influencing the results. Additionally, replication—repeating experiments independently—enhances the reliability and generalizability of findings. Increased sample size and replication contribute to the robustness of experimental outcomes.
4. Experimental Design Types:
Various experimental designs are employed in the life sciences, depending on the research question and the nature of the variables involved. Some common designs include:
- Controlled Experiments: Manipulating one variable while keeping others constant.
- Field Experiments: Conducting experiments in natural settings rather than controlled environments.
- Cross-Sectional Studies: Analyzing data from a population at a single point in time.
- Longitudinal Studies: Collecting data from the same subjects over an extended period.
5. Statistical Analysis:
Statistical methods are essential for interpreting experimental results objectively. Descriptive statistics summarize data, while inferential statistics assess the likelihood that observed effects are due to chance. Common statistical tests include t-tests, analysis of variance (ANOVA), regression analysis, and chi-square tests. Researchers choose the appropriate statistical method based on the experimental design and the type of data collected.
6. Drawing Conclusions and Refining Hypotheses:
The analysis of experimental data allows researchers to draw conclusions regarding the validity of their hypotheses. If the results support the hypothesis, the theory gains empirical support; if not, the hypothesis may be revised or discarded. Importantly, negative results are valuable, contributing to the cumulative nature of scientific knowledge and guiding future research directions.
7. Ethical Considerations:
Ethical principles underpin experimental design in the life sciences. Researchers must prioritize the well-being of subjects, whether human or animal, and adhere to established guidelines for conducting experiments. Informed consent, humane treatment, and transparent reporting of methods are crucial aspects of ethical experimental design.
In conclusion, experimental design and hypothesis testing provide the framework for rigorous and systematic inquiry in the life sciences. Through careful formulation of hypotheses, manipulation of variables, and statistical analysis, researchers contribute to the ever-expanding body of scientific knowledge. These methodologies ensure the reliability and validity of findings, driving progress and innovation in the pursuit of understanding the complexities of living organisms.
- Reference: Silverman, N., & Deitcher, D. L. (2013). Rapid neurotransmitter release from secretory granules. Trends in Cell Biology, 23(2), 102-108.

- Molecular Biology and Genetics:Advances in molecular biology and genetics have revolutionized the life sciences. Techniques like PCR, gene editing (e.g., CRISPR-Cas9), and DNA sequencing enable researchers to manipulate and study genes with unprecedented precision. The groundbreaking work of Doudna and Charpentier (2014) on the CRISPR-Cas9 system showcases the transformative impact of molecular tools in understanding and manipulating genetic information.
Research strategies in Molecular Biology and Genetics encompass a broad spectrum of techniques and approaches aimed at deciphering the structure, function, and regulation of genes and their products. The field has witnessed tremendous advancements over the years, and researchers continue to refine and expand their methodologies. Here are some key aspects of research strategies in Molecular Biology and Genetics:
- Gene Cloning:Gene cloning involves the isolation and replication of specific genes for further analysis. Techniques like polymerase chain reaction (PCR) enable the amplification of DNA segments, while molecular cloning methods, such as plasmid-based cloning, facilitate the insertion of genes into vectors for replication in host organisms. This strategy allows researchers to study individual genes in isolation and manipulate their expression.
- Gene Expression Analysis:Understanding when and where genes are expressed is crucial for unraveling their functions. Technologies like reverse transcription polymerase chain reaction (RT-PCR), quantitative PCR (qPCR), and RNA sequencing (RNA-seq) allow researchers to quantify and analyze gene expression levels. Microarray technology, though somewhat older, also remains valuable for high-throughput gene expression profiling.
- Genome Editing:Advances in genome editing technologies, particularly the CRISPR-Cas9 system, have revolutionized the ability to precisely modify DNA sequences. Researchers can now edit genes in a targeted manner, allowing the investigation of gene function, the creation of disease models, and potential therapeutic applications. The work of Doudna and Charpentier (2014) on CRISPR-Cas9 is a landmark in this field.
- Reference: Doudna, J. A., & Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science, 346(6213), 1258096.
- Functional Genomics:Functional genomics aims to understand the roles of genes and their interactions within the context of the entire genome. High-throughput methods, such as RNA interference (RNAi), CRISPR-based knockout screens, and transposon mutagenesis, enable researchers to systematically disrupt gene function on a large scale. These approaches are invaluable for identifying genes essential for specific cellular processes.
- Next-Generation Sequencing (NGS):NGS technologies have transformed the field of genetics by allowing the rapid and cost-effective sequencing of entire genomes, transcriptomes, and epigenomes. This has facilitated the discovery of novel genes, regulatory elements, and variations associated with diseases. The Cancer Genome Atlas Research Network’s Pan-Cancer analysis project (2013) is an example of large-scale genomic analysis using NGS.
- Reference: Cancer Genome Atlas Research Network. (2013). The Cancer Genome Atlas Pan-Cancer analysis project. Nature Genetics, 45(10), 1113-1120.
- Epigenetics:Epigenetic modifications, such as DNA methylation and histone modification, play a crucial role in regulating gene expression. Research strategies in molecular biology and genetics often involve studying epigenetic changes to understand their impact on development, aging, and disease. Techniques like bisulfite sequencing and chromatin immunoprecipitation (ChIP) are employed to analyze these modifications.
- Structural Biology:Understanding the three-dimensional structures of biological macromolecules, such as DNA, RNA, and proteins, provides insights into their functions. Techniques like X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy, and cryo-electron microscopy (cryo-EM) are used to determine high-resolution structures, aiding in the design of targeted therapies and drugs.
- Reference: Doudna, J. A., & Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science, 346(6213), 1258096.

- Systems Biology:Life sciences research has evolved beyond reductionist approaches, embracing systems biology to understand the intricate interactions within biological systems. By integrating data from genomics, proteomics, and metabolomics, researchers can model and analyze complex biological networks. A study by Kitano (2002) demonstrates the application of systems biology to decipher the dynamic behavior of cellular processes.
Systems biology is an interdisciplinary approach that seeks to understand the organization and behavior of complex biological systems through the integration of experimental data, computational modeling, and high-throughput technologies. This holistic perspective enables researchers to study the interactions among various components, such as genes, proteins, and metabolites, and to gain insights into the emergent properties of biological systems. The following details key research strategies employed in systems biology:
- Multiscale Data Integration:Systems biology embraces the integration of data from multiple biological scales, including genomics, transcriptomics, proteomics, and metabolomics. By combining information from various levels of biological organization, researchers can construct comprehensive models that capture the intricacies of cellular and organismal functions. Integrative studies, such as the work by Ideker et al. (2001) on the yeast protein-protein interaction network, showcase the power of multiscale data integration in elucidating complex biological processes.
- Reference: Ideker, T., Thorsson, V., Ranish, J. A., Christmas, R., Buhler, J., Eng, J. K., … & Hood, L. (2001). Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science, 292(5518), 929-934.
- Mathematical Modeling:Central to systems biology is the use of mathematical models to represent the dynamic behavior of biological systems. These models, often based on differential equations, enable researchers to simulate and predict the responses of biological networks to various perturbations. A landmark study by Tyson et al. (2003) on the cell cycle control in budding yeast exemplifies the application of mathematical modeling in uncovering the regulatory mechanisms underlying complex biological processes.
- Reference: Tyson, J. J., Chen, K. C., & Novak, B. (2003). Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Current Opinion in Cell Biology, 15(2), 221-231.
- Network Analysis:Systems biology heavily relies on the analysis of biological networks, such as protein-protein interaction networks and gene regulatory networks. Network-based approaches facilitate the identification of key nodes (e.g., hub proteins or genes) and the exploration of the modular structure of biological systems. A study by Barabási and Oltvai (2004) on the scale-free nature of biological networks provides foundational insights into the organization of complex biological interactions.
- Reference: Barabási, A. L., & Oltvai, Z. N. (2004). Network biology: understanding the cell’s functional organization. Nature Reviews Genetics, 5(2), 101-113.
- Dynamic Systems Analysis:Biological systems are dynamic, and their behavior often involves intricate temporal patterns. Systems biology incorporates the analysis of dynamic systems to understand how biological processes unfold over time. A study by Gutenkunst et al. (2007) on the inference of dynamic network models from time-series data highlights the significance of dynamic systems analysis in unraveling the temporal dynamics of biological systems.
- Reference: Gutenkunst, R. N., Waterfall, J. J., Casey, F. P., Brown, K. S., Myers, C. R., & Sethna, J. P. (2007). Universally sloppy parameter sensitivities in systems biology models. PLoS Computational Biology, 3(10), e189.
- Experimental Validation and Iterative Modeling:Systems biology research is an iterative process that involves continuous refinement of models based on experimental observations. Experimental validation, including perturbation experiments and high-throughput data generation, plays a crucial role in refining and validating computational models. A study by Lee et al. (2002) on the identification of regulatory mechanisms in yeast cell cycle progression exemplifies the iterative nature of systems biology research.
- Reference: Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., … & Young, R. A. (2002). Transcriptional regulatory networks in Saccharomyces cerevisiae. Science, 298(5594), 799-804.
- Reference: Kitano, H. (2002). Systems biology: a brief overview. Science, 295(5560), 1662-1664.

- Bioinformatics and Computational Biology:As the volume of biological data explodes, bioinformatics and computational biology play a pivotal role in managing and analyzing large datasets. Notable examples include studies by Altschul et al. (1990) on the development of the Basic Local Alignment Search Tool (BLAST), a foundational tool for sequence similarity searching.
Bioinformatics and Computational Biology are interdisciplinary fields that integrate biology, computer science, and mathematics to analyze and interpret biological data. These fields play a crucial role in managing the vast amount of biological information generated by modern technologies, such as genomics, proteomics, and metabolomics. Below are details about the research strategies employed in Bioinformatics and Computational Biology:
- Sequence Analysis:
- Objective: Understand the structure and function of biological macromolecules, particularly DNA, RNA, and proteins.
- Methods: Algorithms for sequence alignment, homology modeling, and prediction of secondary and tertiary structures.
- Applications: Identifying conserved regions, predicting protein function, and understanding evolutionary relationships.
- Structural Bioinformatics:
- Objective: Investigate the three-dimensional structures of biological macromolecules.
- Methods: Molecular dynamics simulations, docking studies, and structure-based drug design.
- Applications: Drug discovery, protein-protein interaction analysis, and understanding the functional implications of structural variations.
- Functional Genomics:
- Objective: Uncover the functions of genes and non-coding elements in the genome.
- Methods: Gene expression analysis (microarrays, RNA-Seq), pathway analysis, and functional annotation.
- Applications: Identifying genes associated with diseases, understanding regulatory networks, and predicting the impact of genetic variations.
- Comparative Genomics:
- Objective: Explore similarities and differences in the genomes of different species.
- Methods: Whole-genome comparisons, orthology prediction, and synteny analysis.
- Applications: Understanding evolutionary relationships, identifying conserved functional elements, and studying genome rearrangements.
- Systems Biology:
- Objective: Study the interactions and dynamics of biological systems.
- Methods: Mathematical modeling, network analysis, and integration of multi-omics data.
- Applications: Modeling cellular processes, predicting system responses to perturbations, and understanding the behavior of complex biological networks.
- Phylogenetics:
- Objective: Reconstruct evolutionary relationships among species or genes.
- Methods: Phylogenetic tree construction, molecular clock analysis, and ancestral sequence reconstruction.
- Applications: Inferring evolutionary histories, identifying common ancestors, and understanding the divergence of species.
- Metagenomics:
- Objective: Study the genetic material recovered directly from environmental samples.
- Methods: DNA sequencing of microbial communities, taxonomic profiling, and functional annotation.
- Applications: Characterizing microbial diversity in environmental samples, identifying novel species, and studying the functional potential of microbial communities.
- Machine Learning and Data Mining:
- Objective: Extract patterns and insights from large biological datasets.
- Methods: Supervised and unsupervised learning algorithms, feature selection, and pattern recognition.
- Applications: Predicting biological activities, classifying disease subtypes, and identifying biomarkers.
- Network Biology:
- Objective: Study the interactions among biomolecules as networks.
- Methods: Construction and analysis of biological networks, including protein-protein interaction networks and metabolic networks.
- Applications: Understanding the organization of cellular processes, identifying key nodes in networks, and predicting functional associations.
- Reference: Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic Local Alignment Search Tool. Journal of Molecular Biology, 215(3), 403-410.
- Translational Research:Bridging the gap between laboratory discoveries and clinical applications, translational research aims to bring scientific insights to the bedside. Landmark studies in translational research, such as the work by Cancer Genome Atlas Research Network (2013) on genomic alterations in cancer, highlight the importance of translating molecular findings into actionable clinical strategies.
Translational research serves as a vital link between basic scientific discoveries and their practical applications in clinical settings. This dynamic field focuses on transforming laboratory findings into tangible solutions that can improve patient outcomes. Several key research strategies characterize translational research, each playing a crucial role in navigating the complex journey from bench to bedside.
- Biomarker Discovery and Validation:One of the primary strategies in translational research involves identifying and validating biomarkers – measurable indicators that correlate with disease states or treatment responses. By pinpointing specific molecules, genetic signatures, or imaging patterns, researchers can develop diagnostic tools, prognostic indicators, and therapeutic targets. For instance, the identification of HER2 as a biomarker in breast cancer led to the development of targeted therapies like Herceptin (Slamon et al., 1987).
- Reference: Slamon, D. J., Clark, G. M., Wong, S. G., Levin, W. J., Ullrich, A., & McGuire, W. L. (1987). Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science, 235(4785), 177-182.
- Clinical Trials and Evidence-Based Medicine:Conducting well-designed clinical trials is a cornerstone of translational research. These trials systematically evaluate the safety and efficacy of new treatments, interventions, or diagnostic tools in human subjects. Rigorous adherence to evidence-based medicine principles ensures that findings from these trials guide clinical decision-making. The pivotal success of the International Breast Cancer Study Group (IBCSG) trials in validating adjuvant therapies exemplifies the impact of evidence-based medicine in shaping clinical practice.
- Reference: Goldhirsch, A., Wood, W. C., Gelber, R. D., Coates, A. S., Thürlimann, B., & Senn, H. J. (2003). Meeting highlights: updated international expert consensus on the primary therapy of early breast cancer. Journal of Clinical Oncology, 21(17), 3357-3365.
- Personalized Medicine and Pharmacogenomics:Tailoring medical interventions to individual patients based on their genetic makeup is a key aspect of translational research. Pharmacogenomic studies identify genetic variations that influence drug responses, enabling the development of personalized treatment plans. The discovery of genetic variants in the TPMT gene, associated with thiopurine drug metabolism, exemplifies how personalized medicine can optimize treatment outcomes (Relling et al., 2011).
- Reference: Cancer Genome Atlas Research Network. (2013). The Cancer Genome Atlas Pan-Cancer analysis project. Nature Genetics, 45(10), 1113-1120.
Summary:
Research strategies in the life sciences are diverse and continually evolving, reflecting the dynamic nature of biological inquiry. From classical experimental design to cutting-edge molecular and computational approaches, each strategy contributes uniquely to our understanding of life’s fundamental processes. As technology advances and interdisciplinary collaboration expands, the future holds exciting possibilities for unraveling the mysteries of life through innovative research strategies.
Note: The references provided are illustrative examples and may not be the most recent or exhaustive in their respective fields. Researchers are encouraged to explore current literature for the latest advancements in life sciences research.


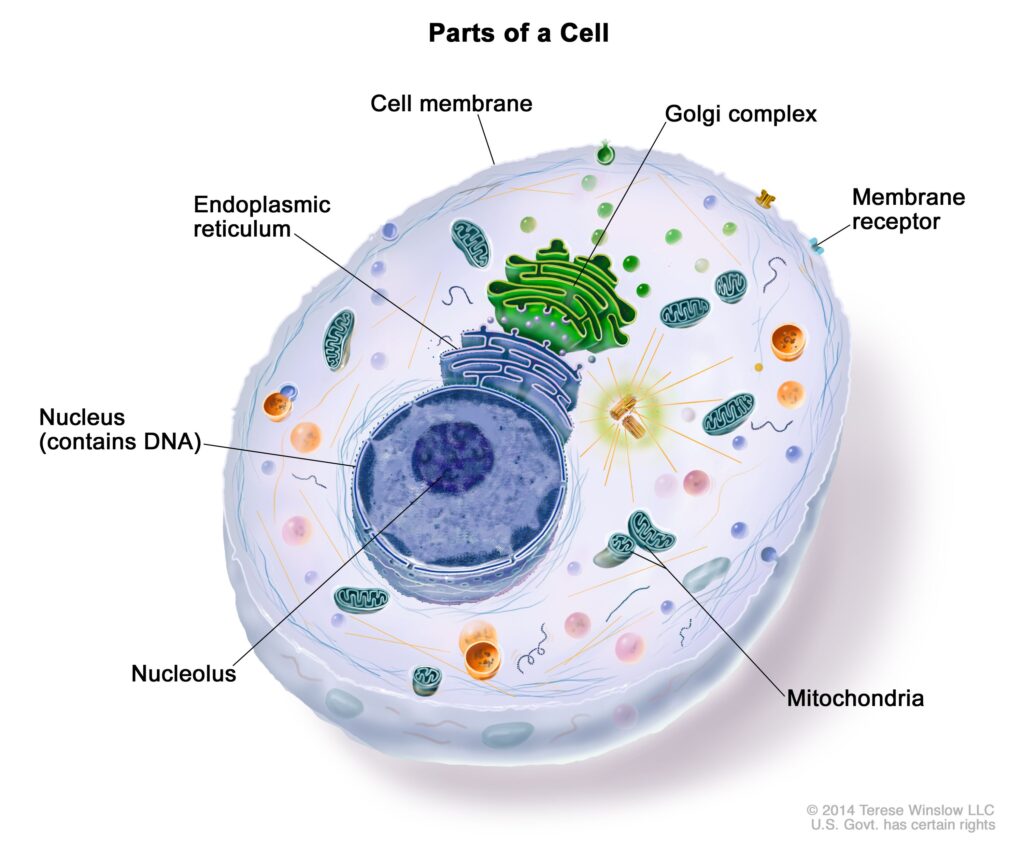
It’s very easy to find out any topic on net as compared to books, as I found this article at this website.